Python Package deadwood Reference¶
|
A base class for |
|
A base class for |
|
A base class for |
|
Deadwood [1] is an anomaly detection algorithm based on Mutual Reachability Minimum Spanning Trees. |
|
Finds the most significant knee/elbow using the Kneedle algorithm [1] with exponential smoothing. |
|
Draws a scatter plot. |
|
Draws a set of disjoint line segments. |
deadwood Python Package
- class deadwood.Deadwood(*, contamination='auto', max_debris_size='auto', M=5, max_contamination=0.5, ema_dt=0.01, metric='l2', quitefastmst_params=None, verbose=False)¶
Deadwood [1] is an anomaly detection algorithm based on Mutual Reachability Minimum Spanning Trees. It trims protruding tree segments and marks small debris as outliers.
More precisely, the use of a mutual reachability distance [3] pulls peripheral points farther away from each other. Tree edges with weights beyond the detected elbow point [2] are removed. All the resulting connected components whose sizes are smaller than a given threshold are deemed anomalous.
Once the spanning tree is determined (\(\Omega(n \log n)\) – \(O(n^2)\)), the Deadwood algorithm runs in \(O(n)\) time. Memory use is also \(O(n)\).
- Parameters:
- M, metric, quitefastmst_params, verbose
see
deadwood.MSTBase- contamination‘auto’ or float, default=’auto’
The estimated (approximate) proportion of outliers in the dataset. If
"auto", the contamination amount will be determined by identifying the most significant elbow point of the curve comprised of increasingly ordered tree edge weights smoothened with an exponential moving average- max_debris_size‘auto’ or int, default=’auto’
The maximal size of leftover connected components that will be considered outliers. If
"auto",sqrt(n_samples)is assumed.- max_contaminationfloat, default=0.5
maximal contamination level assumed when contamination is ‘auto’
- ema_dtfloat, default=0.01
controls the smoothing parameter \(\alpha = 1-\exp(-dt)\) of the exponential moving average (in edge length elbow point detection), \(y_i = \alpha w_i + (1-\alpha) w_{i-1}\), \(y_1 = d_1\)
- Attributes:
- n_samples_, n_features_
see
deadwood.MSTBase- labels_ndarray of shape (n_samples_,)
labels_[i]gives the inlier (1) or outlier (-1) status of the i-th input point.- contamination_float or ndarray of shape (n_clusters_,)
Detected contamination threshold(s) (elbow point(s)). For the actual number of outliers detected, compute
numpy.mean(labels_<0).- max_debris_size_float
Computed max debris size.
- _cut_edges_None or ndarray of shape (n_clusters_-1,)
Indexes of MST edges whose removal forms n_clusters_ connected components (clusters) in which outliers are to be sought. This parameter is usually set by
fitcalled on aMSTClusterer.
References
[2]V. Satopaa, J. Albrecht, D. Irwin, B. Raghavan, Finding a “Kneedle” in a haystack: Detecting knee points in system behavior, In: 31st Intl. Conf. Distributed Computing Systems Workshops, 2011, 166-171, https://doi.org/10.1109/ICDCSW.2011.20
[3]R.J.G.B. Campello, D. Moulavi, J. Sander, Density-based clustering based on hierarchical density estimates, Lecture Notes in Computer Science 7819, 2013, 160-172, https://doi.org/10.1007/978-3-642-37456-2_14
- fit(X, y=None)¶
Detects outliers in a dataset.
- Parameters:
- Xobject
Typically a matrix or a data frame with
n_samplesrows andn_featurescolumns; seedeadwood.MSTBase.fit_predictfor more details.If X is an instance of
deadwood.MSTClusterer(withfitmethod already invoked), e.g.,genieclust.Genieorlumbermark.Lumbermark, then the outliers are sought separately in each detected cluster. Note that this overrides theM,metric, andquitefastmst_paramsparameters (amongst others).If X is None, then the outlier detector is applied on the same MST as that determined by the previous call to
fit. This way we may try out different values of contamination and max_debris_size much more cheaply.- yNone
Ignored.
- Returns:
- selfdeadwood.Deadwood
The object that the method was called on.
Notes
Refer to the labels_ attribute for the result.
As with all distance-based methods (this includes local outlier factor as well), applying data preprocessing and feature engineering techniques (e.g., feature scaling, feature selection, dimensionality reduction) might lead to more meaningful results.
- class deadwood.MSTBase(*, M=0, metric='l2', quitefastmst_params=None, verbose=False)¶
A base class for
genieclust.Genie,lumbermark.Lumbermark,deadwood.Deadwood, and other Euclidean and mutual reachability minimum spanning tree-based clustering and outlier detection algorithms.A Euclidean minimum spanning tree (MST) provides a computationally convenient representation of a dataset: the n points are connected via n-1 shortest segments. Provided that the dataset has been appropriately preprocessed so that the distances between the points are informative, an MST can be applied in outlier detection, clustering, density estimation, dimensionality reduction, and many other topological data analysis tasks.
- Parameters:
- Mint, default=0
Smoothing factor for the mutual reachability distance [1]. M = 0 and M = 1 select the original distance as given by the metric parameter.
- metricstr, default=’l2’
The metric used to compute the linkage.
One of:
"l2"(synonym:"euclidean"; the default),"l1"(synonym:"manhattan","cityblock"),"cosinesimil"(synonym:"cosine"), or"precomputed".For
"precomputed", the X argument to thefitmethod must be a distance vector or a square-form distance matrix; seescipy.spatial.distance.pdist.Determining minimum spanning trees with respect to the Euclidean distance is much faster than with other metrics thanks to the quitefastmst package.
- quitefastmst_paramsdict or None, default=None
Additional parameters to be passed to
quitefastmst.mst_euclidifmetricis"l2".- verbosebool, default=False
Whether to print diagnostic messages and progress information onto
stderr.
- Attributes:
- n_samples_int
The number of points in the dataset.
- n_features_int or None
The number of features in the dataset.
- labels_ndarray of shape(n_samples_, )
Detected cluster labels or outlier flags.
For clustering,
labels_[i]gives the cluster ID of the i-th object (between 0 and n_clusters_ - 1).For outlier detection,
1denotes an inlier.Outliers are labelled
-1.
Notes
If
metricis"l2", then the MST is computed via a call toquitefastmst.mst_euclid. It is efficient in low-dimensional spaces. Otherwise, a general-purpose implementation of the Jarník (Prim/Dijkstra)-like \(O(n^2)\)-time algorithm is called.If M > 0, then the minimum spanning tree is computed with respect to a mutual reachability distance [1]: \(d_M(i,j)=\max\{d(i,j), c_M(i), c_M(j)\}\), where \(d(i,j)\) is an ordinary distance and \(c_M(i)\) is the core distance given by \(d(i,k)\) with \(k\) being \(i\)’s \(M\)-th nearest neighbour (not including self, unlike in [1]). This pulls outliers away from their neighbours.
If the distances are not unique, there might be multiple trees spanning a given graph that meet the minimality property.
- Environment variables:
- OMP_NUM_THREADS
Controls the number of threads used for computing the minimum spanning tree.
References
[1] (1,2,3)R.J.G.B. Campello, D. Moulavi, J. Sander, Density-based clustering based on hierarchical density estimates, Lecture Notes in Computer Science 7819, 2013, 160-172, https://doi.org/10.1007/978-3-642-37456-2_14
[2]M. Gagolewski, A. Cena, M. Bartoszuk, Ł. Brzozowski, Clustering with minimum spanning trees: How good can it be?, Journal of Classification 42, 2025, 90-112, https://doi.org/10.1007/s00357-024-09483-1
[3]M. Gagolewski, quitefastmst, in preparation, 2026, TODO
- fit_predict(X, y=None, **kwargs)¶
Performs cluster or anomaly analysis of a dataset and returns the predicted labels.
Refer to
fitfor a more detailed description.- Parameters:
- Xobject
Typically a matrix or a data frame with
n_samplesrows andn_featurescolumns; see below for more details and options.- yNone
Ignored.
- **kwargsdict
Arguments to be passed to
fit.
- Returns:
- labels_ndarray of shape (n_samples_)
self.labels_ attribute.
Notes
Acceptable X types are as follows.
If X is None, then the MST is not recomputed; the last spanning tree as well as the corresponding M, metric, and quitefastmst_params parameters are selected.
If X is an instance of
MSTBase(or its descendant), all its attributes are copied into the current object (including its MST).For metric is ‘precomputed’, X should either be a distance vector of length
n_samples*(n_samples-1)/2(seescipy.spatial.distance.pdist) or a square distance matrix of shape(n_samples, n_samples)(seescipy.spatial.distance.squareform).Otherwise, X should be real-valued matrix (dense
numpy.ndarrayor an object coercible to) withn_samplesrows andn_featurescolumns.
- class deadwood.MSTClusterer(n_clusters=2, *, M=0, metric='l2', quitefastmst_params=None, verbose=False)¶
A base class for
genieclust.Genie,lumbermark.Lumbermark, and other spanning tree-based clustering algorithms [1].By removing k-1 edges from a spanning tree, we form k connected components which can be conceived as clusters.
- Parameters:
- M, metric, quitefastmst_params, verbose
see
deadwood.MSTBase- n_clustersint
The number of clusters to detect.
- Attributes:
- n_samples_, n_features_
see
deadwood.MSTBase- labels_ndarray of shape (n_samples_,)
Detected cluster labels.
labels_[i]gives the cluster ID of the i-th input point (between 0 and n_clusters_-1).Eventual outliers are labelled
-1.- n_clusters_int
The actual number of clusters detected by the algorithm.
If it is different from the requested one, a warning is generated.
- _cut_edges_ndarray of shape (n_clusters_-1,)
Indexes of the MST edges whose removal forms the n_clusters_ detected connected components (clusters).
References
[1] (1,2)M. Gagolewski, A. Cena, M. Bartoszuk, Ł. Brzozowski, Clustering with minimum spanning trees: How good can it be?, Journal of Classification 42, 2025, 90-112, https://doi.org/10.1007/s00357-024-09483-1
- class deadwood.MSTOutlierDetector(*, M=0, metric='l2', quitefastmst_params=None, verbose=False)¶
A base class for
deadwood.Deadwoodand other spanning tree-based outlier detection algorithms.- Parameters:
- M, metric, quitefastmst_params, verbose
see
deadwood.MSTBase
- Attributes:
- n_samples_, n_features_
see
deadwood.MSTBase- labels_ndarray of shape (n_samples_,)
labels_[i]gives the inlier (1) or outlier (-1) status of the i-th input point.
- deadwood.kneedle_increasing(x, convex=True, dt=0.01)¶
Finds the most significant knee/elbow using the Kneedle algorithm [1] with exponential smoothing.
- Parameters:
- xndarray
data vector (increasing)
- convexbool
whether the data in x are convex-ish (elbow detection) or not (knee lookup)
- dtfloat
controls the smoothing parameter \(\alpha = 1-\exp(-dt)\) of the exponential moving average, \(y_i = \alpha x_i + (1-\alpha) x_{i-1}\), \(y_1 = x_1\).
- Returns:
- indexinteger
the index of the knee/elbow point; 0 if not found
References
[1] (1,2)V. Satopaa, J. Albrecht, D. Irwin, B. Raghavan, Finding a “Kneedle” in a haystack: Detecting knee points in system behavior, In: 31st Intl. Conf. Distributed Computing Systems Workshops, 2011, 166-171, https://doi.org/10.1109/ICDCSW.2011.20
- deadwood.plot_scatter(X, y=None, labels=None, axis=None, title=None, xlabel=None, ylabel=None, xlim=None, ylim=None, asp=None, colors=['#000000', '#DF536B', '#61D04F', '#2297E6', '#28E2E5', '#CD0BBC', '#F5C710', '#1f77b4', '#ff7f0e', '#2ca02c', '#d62728', '#9467bd', '#8c564b', '#e377c2', '#7f7f7f', '#bcbd22', '#17becf', '#1f77b4', '#aec7e8', '#ff7f0e', '#ffbb78', '#2ca02c', '#98df8a', '#d62728', '#ff9896', '#9467bd', '#c5b0d5', '#8c564b', '#c49c94', '#e377c2', '#f7b6d2', '#7f7f7f', '#c7c7c7', '#bcbd22', '#dbdb8d', '#17becf', '#9edae5', '#393b79', '#5254a3', '#6b6ecf', '#9c9ede', '#637939', '#8ca252', '#b5cf6b', '#cedb9c', '#8c6d31', '#bd9e39', '#e7ba52', '#e7cb94', '#843c39', '#ad494a', '#d6616b', '#e7969c', '#7b4173', '#a55194', '#ce6dbd', '#de9ed6', '#3182bd', '#6baed6', '#9ecae1', '#c6dbef', '#e6550d', '#fd8d3c', '#fdae6b', '#fdd0a2', '#31a354', '#74c476', '#a1d99b', '#c7e9c0', '#756bb1', '#9e9ac8', '#bcbddc', '#dadaeb', '#636363', '#969696', '#bdbdbd', '#d9d9d9'], markers=['o', '^', '+', 'x', 'D', 'v', 's', '*', '<', '>', '2'], **kwargs)¶
Draws a scatter plot.
- Parameters:
- Xarray_like
Either a two-column matrix that gives the x and y coordinates of the points or a vector of length n. In the latter case, y must be a vector of length n as well.
- yNone or array_like
The y coordinates of the n points in the case where X is a vector.
- labelsNone or array_like
A vector of n integer labels that correspond to each point in X, that gives its plot style.
- axis, title, xlabel, ylabel, xlim, ylimNone or object
If not None, values passed to matplotlib.pyplot.axis, matplotlib.pyplot.title, etc.
- aspNone or object
See matplotlib.axes.Axes.set_aspect.
- colorslist
List of colours corresponding to different labels.
- markerslist
List of point markers corresponding to different labels.
- **kwargsCollection properties
Further arguments to matplotlib.pyplot.scatter.
Notes
If X is a two-column matrix, then
plot_scatter(X)is equivalent toplot_scatter(X[:,0], X[:,1]).Unlike in matplotlib.pyplot.scatter, for any fixed
j, all pointsX[i,:]such thatlabels[i] == jare always drawn in the same way, no matter themax(labels). In particular, labels 0, 1, 2, and 3 correspond to black, red, green, and blue, respectively.This function was inspired by the
plot()function from the R packagegraphics.Examples
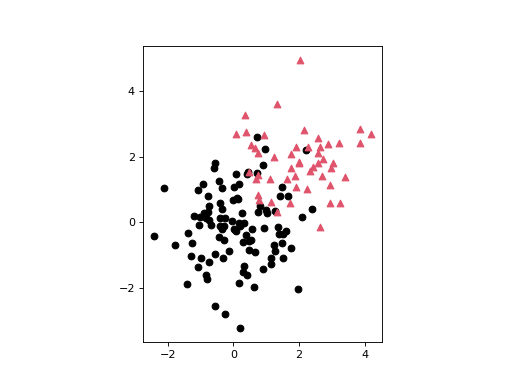
An example scatter plots where each point is assigned one of two distinct labels:
>>> n = np.r_[100, 50] >>> X = np.r_[np.random.randn(n[0], 2), np.random.randn(n[1], 2)+2.0] >>> l = np.repeat([0, 1], n) >>> deadwood.plot_scatter(X, labels=l, asp='equal') >>> plt.show()
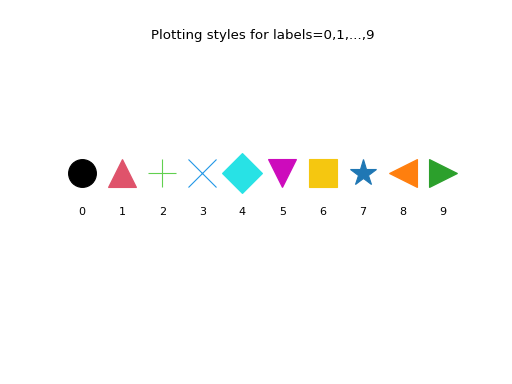
Here are the first 10 plotting styles:
>>> ncol = len(deadwood.colors) >>> nmrk = len(deadwood.markers) >>> mrk_recycled = np.tile( ... deadwood.markers, ... int(np.ceil(ncol/nmrk)))[:ncol] >>> for i in range(10): ... plt.text( ... i, 0, i, horizontalalignment="center") ... plt.plot( ... i, 1, marker=mrk_recycled[i], ... color=deadwood.colors[i], ... markersize=25) >>> plt.title("Plotting styles for labels=0,1,...,9") >>> plt.ylim(-3,4) >>> plt.axis("off") >>> plt.show()
- deadwood.plot_segments(pairs, X, y=None, style='k-', **kwargs)¶
Draws a set of disjoint line segments.
- Parameters:
- pairsarray_like
A two-column matrix that gives the pairs of indexes defining the line segments to draw.
- Xarray_like
Either a two-column matrix that gives the x and y coordinates of the points or a vector of length n. In the latter case, y must be a vector of length n as well.
- yNone or array_like
The y coordinates of the n points in the case where X is a vector.
- style:
See matplotlib.pyplot.plot.
- **kwargsCollection properties
Further arguments to matplotlib.pyplot.plot.
Notes
The function draws a set of disjoint line segments from
(X[pairs[i,0],0], X[pairs[i,0],1])to(X[pairs[i,1],0], X[pairs[i,1],1])for allifrom0topairs.shape[0]-1.matplotlib.pyplot.plot is called only once. Therefore, you can expect it to be pretty fast.
Examples
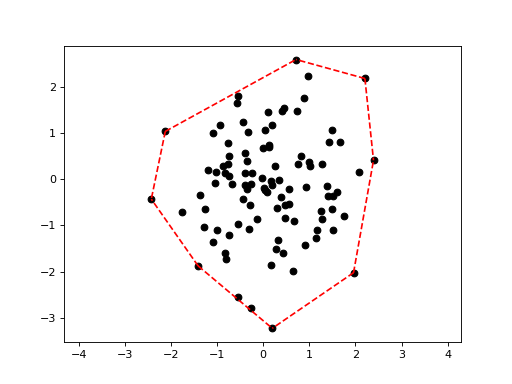
Plotting the convex hull of a point set:
>>> import scipy.spatial >>> X = np.random.randn(100, 2) >>> hull = scipy.spatial.ConvexHull(X) >>> deadwood.plot_scatter(X) >>> deadwood.plot_segments(hull.simplices, X, style="r--") >>> plt.axis("equal") >>> plt.show()
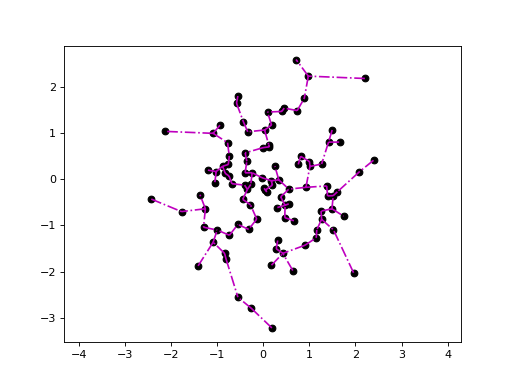
Plotting the Euclidean minimum spanning tree:
>>> import quitefastmst >>> X = np.random.randn(100, 2) >>> mst = quitefastmst.mst_euclid(X) >>> deadwood.plot_scatter(X) >>> deadwood.plot_segments(mst[1], X, style="m-.") >>> plt.axis("equal") >>> plt.show()